Taxonomic Markup and GUIDs
These notes were put together partly in response to discussions with Donat Agosti, but also as part of my experiments with storing ant data in a triple store. The idea is to mark up a taxonomic paper on ants with links to external sources of information (such as names, specimens, images, etc.).
If you want some related inspiration, see Leigh Dodds' post on the scientific paper as a modern palimpsest.
Firstly, I'm going to distinguish between mark up and metadata. I'm going to use mark up to mean tagging a manuscript to identify the relevant bits. For taxonomic literature this is largely after the fact, but for modern journals the article itself is represented in XML, which is then converted to a nice display using XSL. The BMC journals are a good example of this. What I'm interested in is how to mark up an article so that metadata about that article, its contents, and its relationships to other articles can be easily recovered (and output, presumably as RDF). Much of the mark up concerns the structure of a document, which in turn is important for presenting the document (say, in a web browser). I'm interested in just those bits relevant to metadata.
Identifiers
I'm assuming that we have identifiers for the items of interest (i.e., URIs such as URLs, DOIs, Handles, LSIDs). Ideally, there is a way to extract metadata about the object the identifier refers to. LSIDs provide an explicit mechanism for doing this, and CrossRef provides a service to return an XML summary of metadata held for a given DOI.
In my Taxonomic Search Engine I used the Hymenoptera Name Server's SEEK prototype to get metadata about a name, e.g. http://atbi.biosci.ohio-state.edu:210/hymenoptera/nomenclator.seek_demo?id=HNS195070 returns an XML document about the "Google ant", Proceratium google. This document contains these identifiers:
Only the name identifier (HNS195070) has metadata that can be easily accessed in XML, as far as I can see.
How to refer to identifiers
Given the uncertainty about resolving identifiers (i.e., will LSIDs take off), one might adopt the convention used by PubMed and the BMC journals and just include the "local" part of the identifier (see example below), rather than the full blown identifier. Otherwise, a document marked up with the complete identifier will be rendered out of date if the resolution mechanism changes. In English, just use Hymenoptera Namer Server ids, not LSIDs.
Literature cited
Literature is perhaps the least problematic topic, because there are identifiers for many publications (e.g., DOIs), and tools for looking up identifiers for publications (e.g., CrossRef OpenURL, Google Scholar, PubMed, etc.).
BMC uses the following markup for a bibliograpy entry:
Note that individual elements of the item (such as volume, pagination, etc.) are identified, but more importantly, identifiers are provided (in this case from PubMed Central, PubMed, and CrossRef). BMC is better than PLoS in this respect, as PLoS don't embed the identifiers.
Note that the mark up above embeds identifiers, not URLs (for example). URLs are fragile and can break. By just using identifiers, BMC avoids this problem, but it means that the user has to know how to make the identifier actionable.
Marking up literature to this level of detail within even a single paper would be time consuming, but as I've noted elsewhere on iSpecies, tools like ParaCite would make this tractable. ParaCite includes code to generate OpenURL requests, which means finding DOIs would be straightforward.
What's needed?
Tools to extract citations from text and locate identifiers. ParaCite would help. Relying on CrossRef's OpenURL server will be limited to those cases where CrossRef knows about the article (i.e., it has a DOI). It would be useful to have similar tools for searching PubMed, taxon-specific bibliographic databases such as FORMIS, and the Hymenoptera Name Server. By tool I mean a Web API (can be simple as an HTTP GET interface). Google Scholar would also be useful, although there are issues with using it. There is also literature in DSpace repositories, such as the AMNH's wonderful collection of their scientific publications. How do we query this on the fly? In summary, having an OpenURL interface to taxonomic literature would greatly facilitate automated mark up.
Taxonomic names
I subscribe to uBio's view (see doi:10.1080/10635150500541680) that names by themselves are useful and should be indexed. A paper may mention a name with nothing to tell us what taxonomic concept is being used. For example, this is how the Google ant paper describes the site the ants were collected from:
The paper is about ants, and for Discothyrea berlita Fisher, Proceratium avium Brown, Proceratium avioide de Andrade, and Proceratium google Fisher, we have a clear concept of what those names refer to (at a minimum, the specimens listed). For the other names (which include ants and plants), we have little to go on apart from the names. So, every occurrence of a taxonomic name in the document should be flagged. BioOne journals do this already. Any scientific name in the HTML is linked to ITIS. The linking is not intelligent as it is a search, not a link to an identifier (i.e., nobody actually checks that ITIS has the name).
What's needed?
Names in the document should be linked to uBio namebank LSIDs. For ants we could also use the Hymenoptera Name Server.
Taxonomic concepts
If we think of a concept as "what the name means", then this is most relevant to taxonomic papers describing names (e.g., where the author lists specimens, describes features of the taxon), argues that two taxa are synonyms, etc. uBio has a notion of a concept in the sense that a name may exist in multiple classifications, and each combination of name and classification has its own identifier. Hymenoptera has concepts (my understanding is that this is what one sees when one searches the Name Server through the web interface).
Treatment
The taxonomic treatment is probably the same as the taxonomic concept (or perhaps, a treatment can be regarded as a detailed taxonomic concept). In my early experiments I assigned GUIDs to the taxonomic treatment, and used the Dublin Core tag <dcterms:isPartOf> to associate each treatment with the publication. As a quick hack the identifier for each treatment within a paper included the XPath query that would locate that treatment in the larger document, e.g. //tax:treatment[1]. Embedding this much meaning in an identifier is probably not wise, but it meant that given just the identifier, one could extract the treatment from the document.
Authors
The lack of GUIDs for authors is a long standing issue.
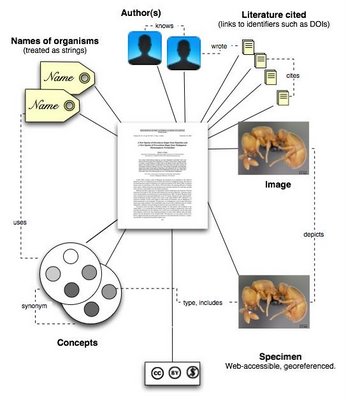
Images
Most taxonomic images probably reside solely within the publication, but some may be stored in external databases (such as AntWeb). In the later case, the mark up should make the link to the external source.
Specimens
If the specimen has an electronic existence, then link to that.

If you want some related inspiration, see Leigh Dodds' post on the scientific paper as a modern palimpsest.
Firstly, I'm going to distinguish between mark up and metadata. I'm going to use mark up to mean tagging a manuscript to identify the relevant bits. For taxonomic literature this is largely after the fact, but for modern journals the article itself is represented in XML, which is then converted to a nice display using XSL. The BMC journals are a good example of this. What I'm interested in is how to mark up an article so that metadata about that article, its contents, and its relationships to other articles can be easily recovered (and output, presumably as RDF). Much of the mark up concerns the structure of a document, which in turn is important for presenting the document (say, in a web browser). I'm interested in just those bits relevant to metadata.
Identifiers
I'm assuming that we have identifiers for the items of interest (i.e., URIs such as URLs, DOIs, Handles, LSIDs). Ideally, there is a way to extract metadata about the object the identifier refers to. LSIDs provide an explicit mechanism for doing this, and CrossRef provides a service to return an XML summary of metadata held for a given DOI.
In my Taxonomic Search Engine I used the Hymenoptera Name Server's SEEK prototype to get metadata about a name, e.g. http://atbi.biosci.ohio-state.edu:210/hymenoptera/nomenclator.seek_demo?id=HNS195070 returns an XML document about the "Google ant", Proceratium google. This document contains these identifiers:
- HNS153344: a taxonomic concept
- HNS195070: a taxonomic name
- pubHNS153344: a publication
Only the name identifier (HNS195070) has metadata that can be easily accessed in XML, as far as I can see.
How to refer to identifiers
Given the uncertainty about resolving identifiers (i.e., will LSIDs take off), one might adopt the convention used by PubMed and the BMC journals and just include the "local" part of the identifier (see example below), rather than the full blown identifier. Otherwise, a document marked up with the complete identifier will be rendered out of date if the resolution mechanism changes. In English, just use Hymenoptera Namer Server ids, not LSIDs.
Literature cited
Literature is perhaps the least problematic topic, because there are identifiers for many publications (e.g., DOIs), and tools for looking up identifiers for publications (e.g., CrossRef OpenURL, Google Scholar, PubMed, etc.).
BMC uses the following markup for a bibliograpy entry:
<bibl id="B21">
<title>
<p>Inter-familial relationships of the shorebirds
(Aves: Charadriiformes) based on nuclear DNA sequence data</p>
</title>
<aug>
<au>
<snm>Ericson</snm>
<fnm>PGP</fnm>
</au>
.
.
.
<au>
<snm>Norman</snm>
<fnm>JA</fnm>
</au>
</aug>
<source>BMC Evol Biol</source>
<pubdate>2003</pubdate>
<volume>3</volume>
<fpage>16</fpage>
<xrefbib>
<pubidlist>
<pubid idtype="pmcid">184354</pubid>
<pubid idtype="pmpid" link="fulltext">12875664</pubid>
<pubid idtype="doi">10.1186/1471-2148-3-16</pubid>
</pubidlist>
</xrefbib>
</bibl>
Note that individual elements of the item (such as volume, pagination, etc.) are identified, but more importantly, identifiers are provided (in this case from PubMed Central, PubMed, and CrossRef). BMC is better than PLoS in this respect, as PLoS don't embed the identifiers.
Note that the mark up above embeds identifiers, not URLs (for example). URLs are fragile and can break. By just using identifiers, BMC avoids this problem, but it means that the user has to know how to make the identifier actionable.
Marking up literature to this level of detail within even a single paper would be time consuming, but as I've noted elsewhere on iSpecies, tools like ParaCite would make this tractable. ParaCite includes code to generate OpenURL requests, which means finding DOIs would be straightforward.
What's needed?
Tools to extract citations from text and locate identifiers. ParaCite would help. Relying on CrossRef's OpenURL server will be limited to those cases where CrossRef knows about the article (i.e., it has a DOI). It would be useful to have similar tools for searching PubMed, taxon-specific bibliographic databases such as FORMIS, and the Hymenoptera Name Server. By tool I mean a Web API (can be simple as an HTTP GET interface). Google Scholar would also be useful, although there are issues with using it. There is also literature in DSpace repositories, such as the AMNH's wonderful collection of their scientific publications. How do we query this on the fly? In summary, having an OpenURL interface to taxonomic literature would greatly facilitate automated mark up.
Taxonomic names
I subscribe to uBio's view (see doi:10.1080/10635150500541680) that names by themselves are useful and should be indexed. A paper may mention a name with nothing to tell us what taxonomic concept is being used. For example, this is how the Google ant paper describes the site the ants were collected from:
Exotic vegetation dominates, most notably a scrub of strawberry guava (Psidium cattleianum) and privet (Ligustrum robustrum)—but grassland and Eucalyptus plantations also occur.
The paper is about ants, and for Discothyrea berlita Fisher, Proceratium avium Brown, Proceratium avioide de Andrade, and Proceratium google Fisher, we have a clear concept of what those names refer to (at a minimum, the specimens listed). For the other names (which include ants and plants), we have little to go on apart from the names. So, every occurrence of a taxonomic name in the document should be flagged. BioOne journals do this already. Any scientific name in the HTML is linked to ITIS. The linking is not intelligent as it is a search, not a link to an identifier (i.e., nobody actually checks that ITIS has the name).
What's needed?
Names in the document should be linked to uBio namebank LSIDs. For ants we could also use the Hymenoptera Name Server.
Taxonomic concepts
If we think of a concept as "what the name means", then this is most relevant to taxonomic papers describing names (e.g., where the author lists specimens, describes features of the taxon), argues that two taxa are synonyms, etc. uBio has a notion of a concept in the sense that a name may exist in multiple classifications, and each combination of name and classification has its own identifier. Hymenoptera has concepts (my understanding is that this is what one sees when one searches the Name Server through the web interface).
Treatment
The taxonomic treatment is probably the same as the taxonomic concept (or perhaps, a treatment can be regarded as a detailed taxonomic concept). In my early experiments I assigned GUIDs to the taxonomic treatment, and used the Dublin Core tag <dcterms:isPartOf> to associate each treatment with the publication. As a quick hack the identifier for each treatment within a paper included the XPath query that would locate that treatment in the larger document, e.g. //tax:treatment[1]. Embedding this much meaning in an identifier is probably not wise, but it meant that given just the identifier, one could extract the treatment from the document.
Authors
The lack of GUIDs for authors is a long standing issue.
Images
Most taxonomic images probably reside solely within the publication, but some may be stored in external databases (such as AntWeb). In the later case, the mark up should make the link to the external source.
Specimens
If the specimen has an electronic existence, then link to that.

2 Comments:
WHY ARENT YOU HAVING A SEARCH THINGY FOR SCIENTIFIC NAMES OF ANIMALS>!?!?!?!?!!?!?!?!?!!?!?!?!?!?!!?!?!?!?!?!?!?!!?!??!?!?!?!?!?!?!?!?!?!?!?!?!?!?!?!?!?!?!?!?!?!?!?!?!?!?!?!!?!?!!?!?!?????????????????????????????!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!??????????????????????????????!!!!!!!!!!!!!!!!!!!!!!!!!!!!???????????????????????????????!!!!!!!!!!!!!!!!!!!!!!!!!!!!!?????????????????????????????!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!????????????????????????????!!!!!!!!!!!!!!!!!!!!!!!!!!!?????????!!!!!!!!!!!!!!!!!!!!!?
By Anonymous, at 11:29 pm
Anonymous, at 11:29 pm
Oh, you mean like iSpecies, or the Taxonomic Search Engine, or uBio perhaps?
By Roderic Page, at 8:36 am
Roderic Page, at 8:36 am
Post a Comment
<< Home