Discovering new things
So, why bother with all this effort to aggregate information into a triple store, I hear you ask? Well, the expectation is that we can learn things we previously didn't know.
For example, consider the ant specimen casent0500379, which is recorded as the source of several sequences in GenBank. These sequences have been obtained by different research groups, and published in different papers.
We can see this immediately if we construct a graph based on the RDF in the triple store. Each node in the graph is a subject. Two nodes, x and y are connected by an edge if there is a triple corresponding to either (?x, ?pred, ?y) or (?y, ?pred, ?x). The neighbourhood of a node is all the nodes adjacent to that node.
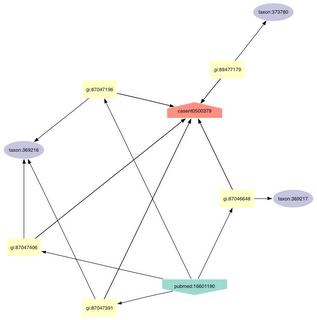
This graph represents the neighbourhood of specimen casent0500379. I've expanded the graph by finding the neighbours of all the neighbours of casent0500379. Note that this graph shows that five sequences have been obtained from this specimen (identified by their "gi" numbers), and those five sequences are associated with three different taxonomic names in GenBank(!).
Two labs have sequenced the 28S ribosomal RNA gene from this specimen, one with accession number DQ353560 (gi:87047406), published by Moreau, et al. in Science (pmid:16601190, doi:10.1126/science.1124891) and one with accession number DQ401020 (gi:89477179), published by Ouellette et al. in MPE (pmid:16630727, doi:10.1016/j.ympev.2006.03.017).
Now, the specimen has not been identified beyond being assigned to the genus Proceratium (which includes the Google ant). These two research groups have given it different informal names, hence GenBank doesn't realise that these two taxa are the same.
In fact there is a third taxon, "Proceratium sp. CSM-2006" for this same specimen, which has been sequenced for wingless.
This is a small example of where aggregating and visualise links between multiple data sources can tell us something we didn't know before.
Postscript
My triple store misses the sixth sequence (yet another 28S rRNA sequence) from this specimen (AY325951, gi:34398469), because the specimen is recorded in the "isolate" field, not the specimen_voucher field:
For example, consider the ant specimen casent0500379, which is recorded as the source of several sequences in GenBank. These sequences have been obtained by different research groups, and published in different papers.
We can see this immediately if we construct a graph based on the RDF in the triple store. Each node in the graph is a subject. Two nodes, x and y are connected by an edge if there is a triple corresponding to either (?x, ?pred, ?y) or (?y, ?pred, ?x). The neighbourhood of a node is all the nodes adjacent to that node.

This graph represents the neighbourhood of specimen casent0500379. I've expanded the graph by finding the neighbours of all the neighbours of casent0500379. Note that this graph shows that five sequences have been obtained from this specimen (identified by their "gi" numbers), and those five sequences are associated with three different taxonomic names in GenBank(!).
Two labs have sequenced the 28S ribosomal RNA gene from this specimen, one with accession number DQ353560 (gi:87047406), published by Moreau, et al. in Science (pmid:16601190, doi:10.1126/science.1124891) and one with accession number DQ401020 (gi:89477179), published by Ouellette et al. in MPE (pmid:16630727, doi:10.1016/j.ympev.2006.03.017).
Now, the specimen has not been identified beyond being assigned to the genus Proceratium (which includes the Google ant). These two research groups have given it different informal names, hence GenBank doesn't realise that these two taxa are the same.
In fact there is a third taxon, "Proceratium sp. CSM-2006" for this same specimen, which has been sequenced for wingless.
This is a small example of where aggregating and visualise links between multiple data sources can tell us something we didn't know before.
Postscript
My triple store misses the sixth sequence (yet another 28S rRNA sequence) from this specimen (AY325951, gi:34398469), because the specimen is recorded in the "isolate" field, not the specimen_voucher field:
source 1..1233
/organism="Proceratium sp. CS-2003-1"
/mol_type="genomic DNA"
/isolate="CASENT0500379"
/db_xref="taxon:237763"
/country="Madagascar"

1 Comments:
The internet is of course very useful for research. But not everyone knows how to properly use the information obtained on the internet. You need to be very careful about research published on the internet, because many publications are made by students who do not use professional services 🖌 that help with writing research papers, and at the same time, these students are also not very knowledgeable about the material they publish. Therefore, using someone's research for your own, you also need to be extremely careful.
By Anonymous, at 2:02 pm
Anonymous, at 2:02 pm
Post a Comment
<< Home