 Vince Smith
Vince Smith has constantly been telling me that for many biologists, "database" means an Excel spreadsheet, and that a big problem is simply getting data into a form that can be used online. Bearing that in mind, and also mindful of how much data is kicking around that isn't in "real" databases, I've been playing with
EditGrid as a tool for adding triples to a triple store. I've
commented on EditGrid elsewhere in the context of collaborative data matrices.
So, here's the situation. In my triple store I have information on ant specimen
INBIOCRI001284215, obtained from AntWeb. Now, AntWeb has no pictures of this specimen. However, John Longino's pages on
Acromyrmex coronatus include pictures of this specimen. How do I get that information into my triple store, without writing RDF?
One approach is to create a spreadsheet with three columns (subject, predicate, object), and create the triples, one per row. Now, I could just do this on my computer using, say, Excel, but that's not nearly cool enough, so I'll use EditGrid. But seriously, I'm going to use EditGrid because:
- You can see it, whereas you can't see a file on my computer
- You and I could collaborate on editing the data in EditGrid
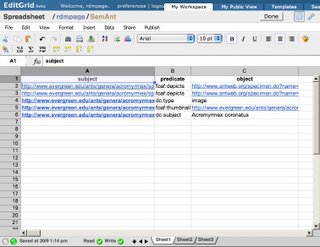
The spreadsheet contains triples, such as these:
In this case the subject and the object are represented by URIs (here they are URLs, but they could also be LSIDs or DOIs). You can see the complete spreadsheet
here. The triples link the picture to the specimen, tell us that http://www.evergreen.edu/ants/genera/acromyrmex/species/coronatus/INBIOCRI001284215_face_orig.jpg is a picture (dc:type image), that the picture has a thumbnail, and is of
Acromyrmex coronatus. Armed with these triples, I can now find a picture of this ant in my triple store.
Fine so far, but how do we get this into the triple store I hear you ask? EditGrid's permalink feature can export the spreadsheet in a range of formats, including XML. So, what I do is grab the
XML, apply a XSL style sheet to convert it to RDF, then import the resulting RDF into the triple store. The key thing is once the data is in the spreadsheet, the rest is trivial. Here's the XSL style sheet. It has limitations, notably the assumption that URIs are URLs.
<?xml version='1.0' encoding='iso-8859-1'?>
<xsl:stylesheet version="1.0" xmlns:xsl="http://www.w3.org/1999/XSL/Transform"
xmlns:rdf="http://www.w3.org/1999/02/22-rdf-syntax-ns#"
xmlns:foaf="http://xmlns.com/foaf/0.1/"
xmlns:dc="http://purl.org/dc/elements/1.1/">
<xsl:output method="xml" version="1.0" encoding="iso-8859-1" indent="yes"/>
<xsl:template match="workbook">
<rdf:RDF>
<xsl:apply-templates select="//row"/>
</rdf:RDF>
</xsl:template>
<xsl:template match="row">
<xsl:if test="@row != '0'">
<xsl:element name="rdf:Description">
<xsl:attribute name="rdf:about">
<xsl:value-of select="cell[1]/@input"/>
</xsl:attribute>
<xsl:variable name="predicate" select="cell[2]/@input"/>
<xsl:variable name="object" select="cell[3]/@input"/>
<xsl:choose>
<xsl:when test="contains($object, 'http://')">
<xsl:element name="{$predicate}">
<xsl:attribute name="rdf:resource">
<xsl:value-of select="$object"/>
</xsl:attribute>
</xsl:element>
</xsl:when>
<xsl:otherwise>
<xsl:element name="{$predicate}">
<xsl:value-of select="$object"/>
</xsl:element>
</xsl:otherwise>
</xsl:choose>
</xsl:element>
</xsl:if>
</xsl:template>
</xsl:stylesheet>
This particular spreadsheet makes some assumptions of the user, namely that they can figure out what is the subject and what is the object, and are comfortable choosing predicates. However, being collaborative, others could help out by editing the spreadsheet. Furthermore, one could create spreadsheets that aren't quite so complicated, and aren't geared towards the developer. For example, one basic source of information I'd like to capture is geographic location, and there is probably a lot more information available in papers than in georeferenced museum collections. Hence, a spreadsheet like this
| observation | lat | long |
| locality | -34.0 | 156.26 |
could be used to capture locality information, and would require minimal effort to convert into RDF. We'd just have to modify the XSL style sheet shown above.
The key point of all of this is that with minimal effort we can capture information that is not in the triple store, and we can make it eas(ish) for people with data to contribute. Given that EditGrid can import Excel files, somebody interested in sharing their data could do the grunt work in Excel on their own computer, then move everything to EditGrid, which makes it accessible to others.
Simple and open wins...



