Automatically growing an ant bibliography
Earlier on iPhylo I'd mentioned the issue of updating a triple store of ants, or indeed, any data base. As an experiment, I've put together a Perl script that can be used to update a data base in Connotea with recent papers on ants. The script makes of a number of web services, and uBio's RSS feeds. It does the following:
Here's a sketch of the process.
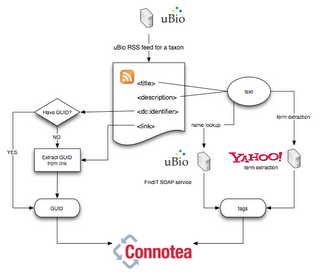
The papers are stored in my semant library. Because it is entirely automated, it could be run regularly (as a cron job, say) to update the library, hence the list of ant papers would grow without any human intervention. At the same, however, users with access to the semant library could manually edit the tags if they feel Yahoo and uBio have missed some relevant terms.
Note also that names recognised by uBio are tagged with LSIDs for the names as well, which means we could resolve those to RDF. In the same way, the Connotea data base itself can serve RDF (here are the ant papers in RDF). Hence, we could easily populate a triple store with metadata about papers and names.
What I like about this script is that it brings together a number of themes.
GUIDs play a key role here. Connotea knows which papers uBio has extracted by using the DOI (or PubMed identifier). Not only does this enable Connotea to know which paper I want, but it uses that identifier to extract metadata about the paper, for example via CrossRef. It also knows whether any other user has already added that paper.
Web services mean that I don't have to reinvent the wheel. If I want to pick out taxonomic names, I use uBio. To extract keywords for tagging, I use Yahoo. To store data, I use Connotea's API.
Tagging makes it easy to add information to a reference.
Social networking through using an open database like Connotea. People can discover other people's libraries through shared papers or shared tags.
RSS pops up at the start and at the end. The whole process starts with a RSS feed (itself an aggregation of numerous journal RSS feeds), and the resulting Connotea data base serves RSS, so others can readily make use of the results.
- Takes an RSS feed for Formicidae from uBio. This feed lists recent papers on ants, as identified using uBio's taxonomic name recognition algorithms.
- Extracts DOIs or PubMed identifiers from the RSS feed. If a DOI isn't found, I see if we can extract one from the <link> tag (typically a URL to the article). uBio does a pretty good job of getting DOIs, but misses some (e.g., for Blackwell and BioOne journals).
- Extracts taxonomic names from the content of the <title> and <description> tags using a SOAP call to uBio's FindIT web service. Ideally, uBio would do this for us, since it has already parsed the journal feed, but for now I do it.
- Uses Yahoo's term extraction web service to extract keywords
- Submit the article GUID (DOI or PubMed id), and the tags to Connotea using the web API.
Here's a sketch of the process.

The papers are stored in my semant library. Because it is entirely automated, it could be run regularly (as a cron job, say) to update the library, hence the list of ant papers would grow without any human intervention. At the same, however, users with access to the semant library could manually edit the tags if they feel Yahoo and uBio have missed some relevant terms.
Note also that names recognised by uBio are tagged with LSIDs for the names as well, which means we could resolve those to RDF. In the same way, the Connotea data base itself can serve RDF (here are the ant papers in RDF). Hence, we could easily populate a triple store with metadata about papers and names.
What I like about this script is that it brings together a number of themes.
GUIDs play a key role here. Connotea knows which papers uBio has extracted by using the DOI (or PubMed identifier). Not only does this enable Connotea to know which paper I want, but it uses that identifier to extract metadata about the paper, for example via CrossRef. It also knows whether any other user has already added that paper.
Web services mean that I don't have to reinvent the wheel. If I want to pick out taxonomic names, I use uBio. To extract keywords for tagging, I use Yahoo. To store data, I use Connotea's API.
Tagging makes it easy to add information to a reference.
Social networking through using an open database like Connotea. People can discover other people's libraries through shared papers or shared tags.
RSS pops up at the start and at the end. The whole process starts with a RSS feed (itself an aggregation of numerous journal RSS feeds), and the resulting Connotea data base serves RSS, so others can readily make use of the results.

62 Comments:
rod
nice stuff
is the "in the news section" in antweb.org actually based on yours, or is this "only" based on UBIO's RSS feed?
This service raises the question again, how to build up a complimentary alert system for gensequences (you have it in updating ants blog), or the one to alert for new ant taxa.
This certainly would fill in some of the users wishes/dreams, especially if the data is provided in a form, it could easily be extracted into their current databases, such as Endnote for literature, exel or access databases...
of course, it's one step closer to a species web page kept automatically updated including addition of all the description of new species.
Donat
By Donat Agosti, at 8:43 am
Donat Agosti, at 8:43 am
Hi everybody!
TermExtractor, my master thesis, is (FREE) online at the
address http://lcl2.di.uniroma1.it.
TermExtractor is a software package for Terminology
Extraction. The software helps a web community to
extract and validate relevant domain terms in their
interest domain, by submitting an archive of
domain-related documents in any format.
TermExtractor extracts terminology consensually
referred in a specific application domain. The
software takes as input a corpus of domain documents,
parses the documents, and extracts a list of
"syntactically plausible" terms (e.g. compounds,
adjective-nouns, etc.).
Documents parsing assigns a greater importance
to terms with text layouts (title, bold, italic,
underlined, etc.). Two entropy-based measures, called
Domain Relevance and Domain Consensus, are then used.
Domain Consensus is used to select only the terms
which are consensually referred throughout the corpus
documents. Domain Relevance to select only the terms
which are relevant to the domain of interest, Domain
Relevance is computed with reference to a set of
contrastive terminologies from different domains.
Finally, extracted terms are further filtered using
Lexical Cohesion, that measures the degree of
association of all the words in a terminological
string. Accept files formats are: txt, pdf, ps, dvi,
tex, doc, rtf, ppt, xls, xml, html/htm, chm, wpd and
also zip archives.
I'd like if you partecipate in the TermExtractor
evaluation task. The result of your evaluation will be
put in a paper (I enclose a draft). Please contact me
if you want to partecipate (this is very important for
me!).
MANY THANKS!!!
--
Francesco Sclano
home page: http://lcl2.di.uniroma1.it/~sclano
msn: francesco_sclano@yahoo.it
skype: francesco978
By Anonymous, at 5:21 pm
Anonymous, at 5:21 pm
good post :)
By Gold Guide for World of Warcraft, at 11:31 pm
Gold Guide for World of Warcraft, at 11:31 pm
Nice blog. I a also ardent player of WOW GOLD. I love this game. Nice posting about wow gold. Thanks
By WOW GOLD, at 9:58 am
WOW GOLD, at 9:58 am
wow great posting!
By BUY WOW GOLD, at 10:51 am
BUY WOW GOLD, at 10:51 am
I am hoping same best effort from you in the future as well. In fact your creative writing skills has inspired me.
SEO services pakistan
By james brownn, at 8:09 pm
james brownn, at 8:09 pm
Nice article you might have carried out below. My business is truly happy to see that. This is the incredibly helpful matter. keep that you're selected it up.Customer Support Services
By Unknown, at 7:24 am
Unknown, at 7:24 am
What a great post with all enjoyably apps visit for more info..:) Buzz Apps
By Unknown, at 5:26 am
Unknown, at 5:26 am
nice
By Jennifer Skelton, at 2:50 pm
Jennifer Skelton, at 2:50 pm
This post give me good info about biblography. We can use this site annotated bibliography service for finding more new topic.
By Jennifer Skelton, at 2:52 pm
Jennifer Skelton, at 2:52 pm
Thanks for sharing such a remarkable post. By following this site rewrite words You'll be able to get detailed info relevant to cheap annotated bibliography service. You can avail this amazing offer. Keep up the good work.
By Anonymous, at 9:35 am
Anonymous, at 9:35 am
You share a great post about biography. It is really needed for me. I have read this information more times. I also found best ideas here about biography writing service. Thanks for this post.
By David Miller, at 2:47 am
David Miller, at 2:47 am
well said! This content is the right way to enhance your knowledge and I like it this post. I want more new updates and keep posting...!
Tableau Training in Chennai
Tableau Course in Chennai
Embedded System Course Chennai
Linux Training in Chennai
Social Media Marketing Courses in Chennai
Pega Training in Chennai
Excel Training in Chennai
Tableau Training in Chennai
Tableau Course in Chennai
By Kayal, at 6:51 am
Kayal, at 6:51 am
What a fabulous post. Would like to thank the admin for sharing this post in our vision.
Spoken English Classes in Chennai
Spoken English Class in Chennai
Spoken English in Chennai
Best Spoken English Classes in Chennai
Best Spoken English Institute in Chennai
IELTS Coaching in Chennai
English Speaking Classes in Mumbai
Spoken English Classes in Anna Nagar
Spoken English Classes in Chennai Anna Nagar
IELTS Classes in Mumbai
By Anbarasan14, at 11:05 am
Anbarasan14, at 11:05 am
Very nice blog, Thank you for providing good information.
Aviation Academy in Chennai
Air hostess training in Chennai
Airport management courses in Chennai
Ground staff training in Chennai
best aviation academy in chennai
cabin crew training institute in chennai
Airline Courses in Chennai
airport ground staff training in chennai
By Anonymous, at 6:55 am
Anonymous, at 6:55 am
the article is nice.most of the important points are there.thankyou for sharing a good one.
JMeter Training in Chennai
JMeter Training Course
JMeter Course
Appium Training in Chennai
Mobile Appium Training in Chennai
Mobile Appium course in Chennai
JMeter Training in Chennai
learn JMeter
By sandeep saxena, at 12:03 pm
sandeep saxena, at 12:03 pm
This blog speaks a great topic which explain very good manner for readers.
web designing and development course training institute in Chennai with placement
PHP MySQL programming developer course training institute in chennai with placement
Magento 2 Developer course training institute in chennai
By Christoper stalin, at 10:57 am
Christoper stalin, at 10:57 am
Nice Presentation and its hopefull words..
if you want a cheap web hosting in web
crm software development company in chennai
erp software development company in chennai
Professional webdesigning company in chennai
best seo company in chennai
By swathima, at 8:16 am
swathima, at 8:16 am
This is the first & best article to make me satisfied by presenting good content. I feel so happy and delighted. Thank you so much for this article... Post office Recruitment 2020 department is issued a new recruitment Notification 2020 For the Post of Postal Assistant and Postman Jobs...
By Anurag Srivastava, at 12:19 pm
Anurag Srivastava, at 12:19 pm
Effective blog with a lot of information. I just Shared you the link below for Courses .They really provide good level of training and Placement,I just Had Android Classes in this institute , Just Check This Link You can get it more information about the Android course. nice apage.
Ai & Artificial Intelligence Course in Chennai
PHP Training in Chennai
Ethical Hacking Course in Chennai Blue Prism Training in Chennai
UiPath Training in Chennai
By subha, at 4:54 pm
subha, at 4:54 pm
thank you for sharing.
Robotic Process Automation (RPA) Training in Chennai | Robotic Process Automation (RPA) Training in anna nagar | Robotic Process Automation (RPA) Training in omr | Robotic Process Automation (RPA) Training in porur | Robotic Process Automation (RPA) Training in tambaram | Robotic Process Automation (RPA) Training in velachery
By sudhan, at 12:19 pm
sudhan, at 12:19 pm
Good. I am really impressed with your writing talents and also with the layout on your weblog. Appreciate, Is this a paid subject matter or did you customize it yourself? Either way keep up the nice quality writing, it is rare to peer a nice weblog like this one nowadays. Thank you, check also virtual edge and 6 Types of Virtual Events You Need to Know for 2021
By Jon Hendo, at 6:00 pm
Jon Hendo, at 6:00 pm
Thank you for your post, I look for such article along time, today i find it finally. this post give me lots of advise it is very useful for me.
AWS Training in Chennai
By Aishwariya, at 5:32 am
Aishwariya, at 5:32 am
Infycle Technologies offers the Best Data training in chennai and is widely known for its excellence in giving the best Data Science Certification course in Chennai. Providing quality software programming training with 100% placement & to build a solid career for every young professional in the software industry is the ultimate aim of Infycle Technologies. Apart from all, the students love the 100% practical training, which is the specialty of Infycle Technologies. To proceed with your career with a solid base, reach Infycle Technologies through 7502633633.
By ram, at 7:16 am
ram, at 7:16 am
Fabulous post... Keep sharing
Best English Coaching Courses in Chennai
Spoken English Courses in Chennai
By chitra, at 11:42 am
chitra, at 11:42 am
Amazing post... Thanks for sharing...
DOT NET Training in Chennai
DOT NET Course in Chennai
By chitra, at 11:08 am
chitra, at 11:08 am
Great post. keep sharing such a worthy information
Digital Marketing Course in Chennai
Best digital marketing course online
Digital Marketing Courses in Bangalore
By Akila, at 12:08 pm
Akila, at 12:08 pm
Great post. keep sharing such a worthy information
Python Training in Chennai
Python Training in Bangalore
By raj, at 11:29 am
raj, at 11:29 am
superb post
By nayar, at 11:44 am
nayar, at 11:44 am
Great post. It is really helpful for me.keep sharing such worthy information..
Web Development courses in Chennai
By priyanka joshna, at 12:42 pm
priyanka joshna, at 12:42 pm
Great post. keep sharing such a worthy information
DevOps course in Chennai
DevOps Course in Bangalore
By Akila, at 10:45 am
Akila, at 10:45 am
Nice info!
RPA course in Chennai
Rpa training online
By Pappu, at 10:31 am
Pappu, at 10:31 am
Infycle Technologies, the best software training institute in Chennai offers the No.1 Python Certification in Chennai for tech professionals. Apart from the Python Course, other courses such as Oracle, Java, Hadoop, Selenium, Android, and iOS Development, Big Data will also be trained with 100% hands-on training. After the completion of training, the students will be sent for placement interviews in the core MNC's. Dial 7502633633 to get more info and a free demo.
By INFYCLE TECHNOLOGIES, at 5:40 pm
INFYCLE TECHNOLOGIES, at 5:40 pm
Reach to the best Python Training institute in Chennai for skyrocketing your career, Infycle Technologies. It is the best Software Training & Placement institute in and around Chennai, that also gives the best placement training for personality tests, interview preparation, and mock interviews for leveling up the candidate's grades to a professional level.
By INFYCLE TECHNOLOGIES, at 5:36 pm
INFYCLE TECHNOLOGIES, at 5:36 pm
Get the best Oracle Training in Chennai from Infycle Technologies, one of the excellent Software Training Institute in Chennai. Great place to study Oracle and we also provide all technical courses like Oracle, Java, Data Science, Big data, AWS, Python, etc. with the best trainers receiving the amazing training for the best career. For more details and demo classes call 7504633633.
By Unknown, at 4:10 pm
Unknown, at 4:10 pm
Norton 360 Crack I desired to thank you for this captivating sensitivity!! I every one taking part in all tiny little bit of it I have you ever bookmarked to test out appendage things you pronounce.
By cyber pc, at 6:47 am
cyber pc, at 6:47 am
Happy Brothers Day Quotes For Younger Brother thanks for the rosy weblog. It turned into without a doubt useful for me. i am happy i found this blog. thanks for sharing bearing in mind us,I too constantly examine some thing secondary out of your pronounce.
By Back linker, at 3:07 am
Back linker, at 3:07 am
Very nice blog.
Egg rate
Barwala egg rate today
Hyderabad egg rate today
Kolkata egg rate today
ajmer egg rate today
namakkal egg rate today
mumbai egg rate today
By Barwalaeggrate, at 1:22 pm
Barwalaeggrate, at 1:22 pm
Great blog with good content and thanks for sharing
Also Visit:
DevOps Training in Chennai |
DevOps Online Course |
DevOps Training in Coimbatore
By Rosy S, at 7:42 am
Rosy S, at 7:42 am
Machines may learn from experience, adapt to new inputs, and carry out activities similar to those performed by humans, thanks to artificial intelligence (AI). Join the Artificial Intelligence Course in Chennai at FITA Academy to learn more about AI.
Artificial Intelligence Course in Chennai
Best AI Courses Online
Artificial Intelligence Course In Bangalore
By priya, at 6:46 am
priya, at 6:46 am
Your self-confidence will grow and any doubts you have will be as cleared as result of your speaking and listening to others in English. There are numerous different ways to speak the language that can help you fast advance your English skills in addition to giving you a confidence boost. Every skill you've developed while learning the language up to this point must be immediately applied while communicating with an English speaker. Any grammatical and vocabulary errors can be found by speaking English aloud. Enroll in Spoken English Classes in Chennai for training; experts at. FITA Academy guide you in a professional way.
By praveen, at 12:01 pm
praveen, at 12:01 pm
Very nice post. Thank you for sharing with us.
Thirumana porutham
Rasi Natchathiram
Numerology Calculator in Tamil
By bharanidevi, at 2:03 pm
bharanidevi, at 2:03 pm
Very informative blog
Hanuman Chalisa Lyrics pdf
Hanuman Chalisa Tamil pdf
Hanuman Chalisa English Pdf
Hanuman Chalisa Hindi Pdf
Hanuman Chalisa Bengali Pdf
Hanuman Chalisa Malayalam Pdf
Hanuman Chalisa Gujarati Pdf
Hanuman Chalisa Kannada Pdf
By hanumanchalisa, at 6:53 am
hanumanchalisa, at 6:53 am
Nice Blog
Oracle Certification in Chennai
Oracle Certification Courses Online
Oracle Course in Coimbatore
By Viswa, at 10:09 am
Viswa, at 10:09 am
In the European Union, German is the most frequently spoken language, surpassing Spanish, French, and even English. In addition to being one of the official languages in Switzerland and Luxembourg, it is the national tongue of Germany.
German Classes in Chennai
German Classes In Bangalore
German Classes in Coimbatore
By priya, at 6:40 am
priya, at 6:40 am
To know more about JMeter join Jmeter Training In Chennai at FITA Academy.
Jmeter Training In Chennai
Jmeter Certification
Jmeter Training in Bangalore
By priya, at 9:53 am
priya, at 9:53 am
Thanks for this blog, it is for informative.
Bnp Paribas Careers
Bnp Paribas Jobs For Freshers
By Viji, at 12:25 pm
Viji, at 12:25 pm
Great Content..Thanks for Sharing
Artificial Intelligence Course in Chennai
Best AI Courses Online
Artificial Intelligence Course In Bangalore
By Nithiya, at 1:27 pm
Nithiya, at 1:27 pm
very nice post thanks fo sharing this post
Hanuman Chalisa Lyrics Pdf
Hanuman Chalisa Tamil Pdf
Hanuman Chalisa English Pdf
Hanuman Chalisa Telugu Pdf
Hanuman Chalisa malayalam Lyrics Pdf
Hanuman Chalisa Bengali Pdf
Hanuman Chalisa Gujarati pdf
Hanuman Chalisa Kannada PDF
By hanumanchalisa, at 9:52 am
hanumanchalisa, at 9:52 am
Thanks for sharing valuable information, keep posting Software Testing Classes in Pune
Best It Training Provider
By Anonymous, at 4:57 am
Anonymous, at 4:57 am
React Native was developed to make the development of mobile apps easier after ReactJS. The answer is straightforward: life is made much simpler if you can create an app once in JavaScript and release it to both Android and iOS.Using JavaScript, you can create native mobile apps with the React Native framework. Typically, Java (for Android) and Swift/Obj-C are required for developing mobile apps (for iOS). With React Native, you can create fully functional apps for both platforms in less time and with only one code language. To learn more about react native, join React Native Training In Chennai at FITA Academy.
React Native Training In Chennai
By Divya, at 12:42 pm
Divya, at 12:42 pm
Thanks for sharing this post very nice post
Sai Satcharitra Pdf
Sai Satcharitra Telugu Pdf
By saipdf, at 5:02 pm
saipdf, at 5:02 pm
Thanks for sharing valuable information, keep us posted. Python Classes in Pune
Best IT Training Provider
By Anonymous, at 8:17 am
Anonymous, at 8:17 am
Thanks for sharing post . java training in pune
By shubhi pandey, at 8:21 am
shubhi pandey, at 8:21 am
thanks for valuable info
gcp training in hyderabad
By GCP MASTERS, at 7:46 am
GCP MASTERS, at 7:46 am
This comment has been removed by the author.
By CarlosOP, at 10:36 pm
CarlosOP, at 10:36 pm
Your blog post was a joy to read! The way you National egg rate explained everything was clear and interesting, making the topic easy to understand.
By CarlosOP, at 11:15 pm
CarlosOP, at 11:15 pm
Great Articles...Thanks for sharing your articles, Keep doing this.
scrum-master-training-in-hyderabad
By kosmik, at 11:36 am
kosmik, at 11:36 am
Excellent article.
Experiencing Sai Baba's blessings
The timeless wisdom of Sai Baba
By delphinemary, at 4:59 pm
delphinemary, at 4:59 pm
A strong academic foundation is often built during the higher classes, where students are guided to develop not only knowledge but also life skills. At this stage, schools focus on nurturing critical thinking, creativity, and discipline, ensuring learners are prepared for future academic and professional challenges. The environment is designed to encourage curiosity and holistic growth.
For parents seeking a trusted institution that emphasizes both academic excellence and character development, choosing the right senior secondary school in Pondicherry becomes essential. Such schools provide well-structured programs, modern facilities, and supportive mentorship that help students achieve success in every sphere of life.
By Amalorpavam, at 11:21 am
Amalorpavam, at 11:21 am
This comment has been removed by the author.
By المثالي كلين, at 1:46 pm
المثالي كلين, at 1:46 pm
بالتوفيق اعمال مميزة شارك اعمالنا تُعد الخيار الأمثل لكل من يسعى لتحسين جودة حياته المنزلية من خلال خدمات تنظيف شاملة تسليك مجاري محترف، ومكافحة فعالة للحشرات شارك اعمال شركة تسليك مجاري بسيهات التزامها بالجودة والاحترافية يجعلها تبرز بين الشركات الأخرى ويضعها في مقدمة قائمة الخيارات لكل من يرغب في تحويل منزله إلى بيئة مثالية .
By المثالي كلين, at 2:24 pm
المثالي كلين, at 2:24 pm
Post a Comment
<< Home